 |
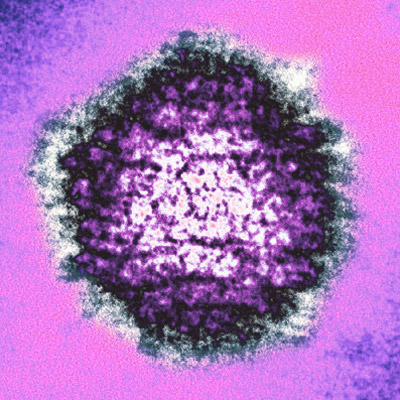
| Herpes Simplex Virus (from AJ Cann) |
Wednesday, November 14, 2012
A New Vaccine Strategy
Saturday, September 22, 2012
For the love of all things scientific, can we please build better databases??
Many scientists work with animal models to learn about human diseases. While most are far from perfect models, we owe a lot of medical advances to them. I work with an animal model for HIV/AIDS and we use a database that was developed onsite to track clinical information about our animals. For example, when one of my animals is bled or has a clinical procedure, the veterinarians and technicians will input stats like blood pressure and heart rate, as well as make notes of any unusual signs or symtoms, like if the blood clotted earlier than usual. Now I need this data from the database so I can make charts of the information about the animals in my study. Unfortunately, this database does not allow for exporting data to other programs and doesn't even let you select data to copy and paste into another program. So when I want a chart that plots the animals' weights from birth to present, I can ask the database for a report and I can tell it to put all of the weight records in chronological order and it will give me two columns of information, date of record and weight. However (!), I then have to manually enter each datapoint into excel or graphpad in order to get my plot. It's an incredibly tedious task and a huge waste of time but that's not the biggest shame of having a primitive database system. The thing about the system that gets me is that the software could easily be used to correlate information across data fields. This feature is something I've long lamented that the medical community lacks. Though the things holding progress back in that sector are mostly privacy issues but we don't have privacy issues in animal research!
So for each animal we have a wealth of biological data sitting in this database that we can't easily analyze and then each department has huge amounts of their own data for subsets of the animals. For example, the genetics department has looked at the genotypes of the animals for genes they are interested in and the virology and immunology department has information about viral status and immune responses and all sorts of other interesting data. but they are all on separate computers and in separate programs. If we could have one central database where every researcher enters information about each animal they study, then we could easily correlate things like genotype to viral load or immune status. but wait! If this database could automatically look for correlations across fields, then we wouldn't even have to wait until some idea occurred to a scientist! The computer could automatically report any correlations and we could look further into them to weed out the false positives. It would also be a boon for epidemiology within the colonies. If there were reports of symptoms spreading through a group of animals, the technicians would make a note of it and we could look for clusters of notes about runny noses or coughing...
Now I have a few friends who can program and are big supporters of research. I have no idea how big of a task it would be to write such a program but if we could find enough programers who also dig on science and want to help scientific research, I wonder if this program could be developed on a volunteer basis. I mean with all of the open source software that people write and improve on everyday, surely there are the means to make this incredibly useful software, right?? What's more, if I could set up something like a kickstarter to get a pool of money to pay some programers, would there be enough interest from the public to make that pool big enough? And could this program evolve into a functional database for human biological information? That would be incredible! Though the hurdles there would be substantial and the bureaucracy perhaps too daunting for me. At any rate, I'm going to toy with these ideas for a while.
So for each animal we have a wealth of biological data sitting in this database that we can't easily analyze and then each department has huge amounts of their own data for subsets of the animals. For example, the genetics department has looked at the genotypes of the animals for genes they are interested in and the virology and immunology department has information about viral status and immune responses and all sorts of other interesting data. but they are all on separate computers and in separate programs. If we could have one central database where every researcher enters information about each animal they study, then we could easily correlate things like genotype to viral load or immune status. but wait! If this database could automatically look for correlations across fields, then we wouldn't even have to wait until some idea occurred to a scientist! The computer could automatically report any correlations and we could look further into them to weed out the false positives. It would also be a boon for epidemiology within the colonies. If there were reports of symptoms spreading through a group of animals, the technicians would make a note of it and we could look for clusters of notes about runny noses or coughing...
Now I have a few friends who can program and are big supporters of research. I have no idea how big of a task it would be to write such a program but if we could find enough programers who also dig on science and want to help scientific research, I wonder if this program could be developed on a volunteer basis. I mean with all of the open source software that people write and improve on everyday, surely there are the means to make this incredibly useful software, right?? What's more, if I could set up something like a kickstarter to get a pool of money to pay some programers, would there be enough interest from the public to make that pool big enough? And could this program evolve into a functional database for human biological information? That would be incredible! Though the hurdles there would be substantial and the bureaucracy perhaps too daunting for me. At any rate, I'm going to toy with these ideas for a while.
Thursday, September 20, 2012
Chronic Fatigue Syndrome and XMRV
 |
| Photo by Phil Myers |
The history of this research is also pretty interesting and has some good ol' human drama in it. Originally, there were two studies that found evidence of XMRV (Lombardi et al 2009) or another mouse virus, pMLV (Lo et al. 2010) in samples from patients with CFS. After these were published, other researchers were not able to replicate the results and eventually the link was dismissed by many scientists. One of the lead scientists in the 2009 study that reported a link would not share her data with other scientists (red flag!) and was eventually fired and briefly arrested for stealing the lab notebooks with the data. She returned the notebooks, though I'm not sure what was found within the data. At any rate, she and the other scientist who found a link between a virus and CFS were both involved in this new study that found no link in a large cohort, which is probably the best possible end to the dispute. It's great that they were able to put their attachment to their original findings aside and work together to figure out what's really going on. I find the human drama part of the story interesting because, as a young scientist, I'm sometimes surprised by how often egos and politics get in the way of scientific progress. Though I'm not as naive about it as I was just a few years ago. At least it makes things more interesting.
Thursday, October 20, 2011
Drug Resistance in Malaria
 |
| P. falciparum gametocytes on a blood smear |
The department seminar last week was given by Dr. Timothy Anderson, a scientist working right here in San Antonio at The Texas Biomedical Research Institute. His lab is looking at genes responsible for drug resistance in the parasite that causes malaria and tracking these mutations as they spread through endemic regions. Drug resistance is an increasing problem for endemic areas where the drugs that are currently used to treat malaria are becoming less effective.

One gene in particular, pfmdr-1, has been well studied and shown to play a role in drug resistance. Copy number variations, as well as single nucleotide polymorphisms (SNPs) in this gene can lead to drug resistance. Dr. Anderson's lab genotyped 160 infections from Malawi to determine the rate of CNVs and SNPs associated with resistance*. All the parasites had a single copy of pfmdr-1. Although no CNVs were seen in this study, this finding is important for setting a baseline of CNV prevalence for future surveillance studies to reference. It was also found that 34% of the parasites had variations at 4 of the 5 SNP sites studied. After determining the prevalence of these mutations in the population, they looked at susceptibility to various anti-malarial drugs and found that several of these genotypes were associated with increased resistance to the drugs tested.
In order to develop effective strategies for drug development and delivery it's important to understand the underlying mechanisms of resistance and how resistance moves through a population. This study and studies like it are critical for understanding how to minimize the spread of drug resistance and for the implementation of smart drug policies that save lives.
*Nkhoma et al. Parasites bearing a single copy of the multi-drug resistance gene (pfmdr-1) with wild-type SNPs predominate amongst Plasmodium falciparum isolates from Malawi. Acta Trop (2009) vol. 111 (1) pp. 78-81
Tuesday, October 18, 2011
First seminar of the semester!
(this post is from September 1, 2011 but I didn't get it published until today... ah procrastination)
So this is the final year of my graduate studies and I'd like to write at least something about each seminar I attend. The first seminar for the Microbiology and Immunology department was today. Woot! It is presented by Duncan Wilson from Albert Einstein College of Medicine. The title was "Studying Herpes Simplex Virus Microtubule-traffic using an in vitro System."
 Dr. Wilson is known for determining where the virus gets its final membrane, which is not as straight forward a question as it might seem. That was more than 20 years ago and today he is working on answering questions regarding how the virus gets around inside a cell and from one cell to another within the nervous system.
Dr. Wilson is known for determining where the virus gets its final membrane, which is not as straight forward a question as it might seem. That was more than 20 years ago and today he is working on answering questions regarding how the virus gets around inside a cell and from one cell to another within the nervous system.
Herpes simplex virus (HSV) is in a family of viruses that are enveloped and use linear double-stranded DNA, which is encased in a capsid. The capsid is surrounded by a tegument of unknown function and the whole thing is wrapped in a membrane derived (as Dr. Wilson's lab showed) from the trans-golgi network (image above).
 HSV infects neurons and in order to move to the nuclear pore for replication, as well as to get from one neuron to the next, they use motor proteins that the cells normally use for trafficking cellular molecules along microtubules (left image). It isn't currently clear whether the whole virus travels along microtubules or if just the capsid with the DNA traverses the cells. This has lead to two models being hypothesized, the separate model (capsid without envelope) and the married model (complete virus). This is an important question not only for the sake of understanding HSV biology but has implications for drug development strategies.
HSV infects neurons and in order to move to the nuclear pore for replication, as well as to get from one neuron to the next, they use motor proteins that the cells normally use for trafficking cellular molecules along microtubules (left image). It isn't currently clear whether the whole virus travels along microtubules or if just the capsid with the DNA traverses the cells. This has lead to two models being hypothesized, the separate model (capsid without envelope) and the married model (complete virus). This is an important question not only for the sake of understanding HSV biology but has implications for drug development strategies.
In order to address this question many researchers are looking at the proteins involved in the process to get an idea of what is going on. Dr. Wilson's lab has identified a large tegument protein that is involved in anterograde trafficking. Because of it's location in the tegument and not on the envelope of the virus, this seems to support the hypothesis that the capsid is responsible for trafficking of the virus. However, some of the electron microscopy data show a capsid that is partially enveloped, suggesting that perhaps a model that is a bit of a hybrid of the separate and married models is actually represent what is going on in the cells.
So this is the final year of my graduate studies and I'd like to write at least something about each seminar I attend. The first seminar for the Microbiology and Immunology department was today. Woot! It is presented by Duncan Wilson from Albert Einstein College of Medicine. The title was "Studying Herpes Simplex Virus Microtubule-traffic using an in vitro System."
 Dr. Wilson is known for determining where the virus gets its final membrane, which is not as straight forward a question as it might seem. That was more than 20 years ago and today he is working on answering questions regarding how the virus gets around inside a cell and from one cell to another within the nervous system.
Dr. Wilson is known for determining where the virus gets its final membrane, which is not as straight forward a question as it might seem. That was more than 20 years ago and today he is working on answering questions regarding how the virus gets around inside a cell and from one cell to another within the nervous system.Herpes simplex virus (HSV) is in a family of viruses that are enveloped and use linear double-stranded DNA, which is encased in a capsid. The capsid is surrounded by a tegument of unknown function and the whole thing is wrapped in a membrane derived (as Dr. Wilson's lab showed) from the trans-golgi network (image above).
 HSV infects neurons and in order to move to the nuclear pore for replication, as well as to get from one neuron to the next, they use motor proteins that the cells normally use for trafficking cellular molecules along microtubules (left image). It isn't currently clear whether the whole virus travels along microtubules or if just the capsid with the DNA traverses the cells. This has lead to two models being hypothesized, the separate model (capsid without envelope) and the married model (complete virus). This is an important question not only for the sake of understanding HSV biology but has implications for drug development strategies.
HSV infects neurons and in order to move to the nuclear pore for replication, as well as to get from one neuron to the next, they use motor proteins that the cells normally use for trafficking cellular molecules along microtubules (left image). It isn't currently clear whether the whole virus travels along microtubules or if just the capsid with the DNA traverses the cells. This has lead to two models being hypothesized, the separate model (capsid without envelope) and the married model (complete virus). This is an important question not only for the sake of understanding HSV biology but has implications for drug development strategies. In order to address this question many researchers are looking at the proteins involved in the process to get an idea of what is going on. Dr. Wilson's lab has identified a large tegument protein that is involved in anterograde trafficking. Because of it's location in the tegument and not on the envelope of the virus, this seems to support the hypothesis that the capsid is responsible for trafficking of the virus. However, some of the electron microscopy data show a capsid that is partially enveloped, suggesting that perhaps a model that is a bit of a hybrid of the separate and married models is actually represent what is going on in the cells.
Tuesday, July 5, 2011
Prions!
I study infectious diseases because I find them so incredibly interesting and varied. Even though my work focuses on viruses, I have mad love for all infectious agents and prions are some of the most interesting. I mean, infectious proteins? How interesting is that? These disease are caused by an altered form of a protein that can cause the native form to become altered. In mammals, a protein named PrP (prion protein) is responsible for these diseases. The altered form is called PrPSc (for scrapie, the name of the prion disease found in sheep) and the native form PrPC (cellular). The altered form of the protein is resistant to degradation and form aggregates resulting in plaques in the brain. These diseases fall into three categories based on mode of transmission: sporadic (caused by a random mutation in the PrP gene), familial (inheritance of mutant PrP) or transmissible (infectious).
You likely know about the widely publicized infectious prion diseases like mad-cow disease or Creutzfeldt–Jakob disease but my favorite prion disease is actually not infectious. It is a hereditary form called fatal familial insomnia (FFS). That's right, insomnia. If you have the misfortune of inheriting this disease, you will die of insomnia. It sounds so so terrible. Luckily, it is exceedingly rare.
You likely know about the widely publicized infectious prion diseases like mad-cow disease or Creutzfeldt–Jakob disease but my favorite prion disease is actually not infectious. It is a hereditary form called fatal familial insomnia (FFS). That's right, insomnia. If you have the misfortune of inheriting this disease, you will die of insomnia. It sounds so so terrible. Luckily, it is exceedingly rare.
Thursday, April 21, 2011
HIV in the gut
Yesterday I attended a seminar by Dr. Ronald Veazy, a scientist in the Pathology Department at Tulane National Primate Research Center. His research focuses on the effects of HIV infection on the gut. His lab works with the animal model of HIV, which is SIV infection of Asian macaques.
For a little background on this model, HIV originated from an SIV (simian immunodeficiency virus) that naturally infects chimpanzees (though now there is some debate on how "natural" this infection is. more on this later...). SIV doesn't result in the same disease course in chimps because they have had some time to adapt to the virus. So while they are not able to get rid of the infection, they are able to control the virus to some extent and death from SIV in chimps seems to be pretty rare.
However, Asian primates are not naturally infected with SIV and when infected display a disease course much like AIDS in humans. This was discovered when a captive rhesus macaque was accidentally infected with an SIV from a sooty mangabee (SIVsm), which gave rise to the virus that is currently used in the animal model, which we now call SIVmac.
Now back to Dr. Veazey's work. Following the discovery of HIV/AIDS, it was thought that the pathogenesis of the virus was predominantly a result of depletion of immune cells called CD4 positive T cells or helper T cells in the blood (from now on CD4+ cells). These cells play a central role in the immune response to invaders and are the target cells of HIV/SIV. The virus uses the CD4 molecule on the surface of these cells (along with a co-receptor) to enter the cells and turn them into little virus factories. These cells can be isolated from the blood and counted to determine the state of the immune system during HIV infection. However, these cells are also found in large numbers in the gut, in what we call gut-associated lymphoid tissue or GALT. The depletion of these cells in the gut was not appreciated as a source of pathogenesis until around 1998 when a group of scientists, including Dr. Veazey, discovered that in macaques these cells are quickly depleted following infection and, unlike the cells in the circulation, these populations of CD4+ cells do not recover after this initial depletion. Tracking these populations in humans was difficult (gut biopsies had to be taken, which is something to avoid in people with impaired immune systems). So there was no evidence of this in humans and it was thought to be a peculiarity of the macaque model until 2004 when it was shown to occur in humans with HIV as well. In the mean time, the co-receptor for HIV was discovered (a molecule called CCR5) and was shown to have high expression in CD4+ cells in the gut. So we now know that T cell depletion in the gut plays a central role in HIV pathogenesis, though the exact mechanism is still under some debate. It was initially assumed that the decrease in CD4+ T cells itself directly leads to immunodeficiency. However, it now seems that the depletion of CD4+ cells in the gut leads to gut "leakiness" that in turn results in systemic inflammation at levels too high to sustain, thus exhausting the immune system and leading to immunodeficiency. Regardless of whether this depletion is the key to disease progression or is just one of many factors that leads to AIDS, Dr. Veazey's research has been incredibly important in the discovery of an important factor in HIV/AIDS pathogenesis.
For a little background on this model, HIV originated from an SIV (simian immunodeficiency virus) that naturally infects chimpanzees (though now there is some debate on how "natural" this infection is. more on this later...). SIV doesn't result in the same disease course in chimps because they have had some time to adapt to the virus. So while they are not able to get rid of the infection, they are able to control the virus to some extent and death from SIV in chimps seems to be pretty rare.
However, Asian primates are not naturally infected with SIV and when infected display a disease course much like AIDS in humans. This was discovered when a captive rhesus macaque was accidentally infected with an SIV from a sooty mangabee (SIVsm), which gave rise to the virus that is currently used in the animal model, which we now call SIVmac.
Now back to Dr. Veazey's work. Following the discovery of HIV/AIDS, it was thought that the pathogenesis of the virus was predominantly a result of depletion of immune cells called CD4 positive T cells or helper T cells in the blood (from now on CD4+ cells). These cells play a central role in the immune response to invaders and are the target cells of HIV/SIV. The virus uses the CD4 molecule on the surface of these cells (along with a co-receptor) to enter the cells and turn them into little virus factories. These cells can be isolated from the blood and counted to determine the state of the immune system during HIV infection. However, these cells are also found in large numbers in the gut, in what we call gut-associated lymphoid tissue or GALT. The depletion of these cells in the gut was not appreciated as a source of pathogenesis until around 1998 when a group of scientists, including Dr. Veazey, discovered that in macaques these cells are quickly depleted following infection and, unlike the cells in the circulation, these populations of CD4+ cells do not recover after this initial depletion. Tracking these populations in humans was difficult (gut biopsies had to be taken, which is something to avoid in people with impaired immune systems). So there was no evidence of this in humans and it was thought to be a peculiarity of the macaque model until 2004 when it was shown to occur in humans with HIV as well. In the mean time, the co-receptor for HIV was discovered (a molecule called CCR5) and was shown to have high expression in CD4+ cells in the gut. So we now know that T cell depletion in the gut plays a central role in HIV pathogenesis, though the exact mechanism is still under some debate. It was initially assumed that the decrease in CD4+ T cells itself directly leads to immunodeficiency. However, it now seems that the depletion of CD4+ cells in the gut leads to gut "leakiness" that in turn results in systemic inflammation at levels too high to sustain, thus exhausting the immune system and leading to immunodeficiency. Regardless of whether this depletion is the key to disease progression or is just one of many factors that leads to AIDS, Dr. Veazey's research has been incredibly important in the discovery of an important factor in HIV/AIDS pathogenesis.
Subscribe to:
Posts (Atom)
